Principal Investigator
:
Dr hab. Marcin Nowotny
International Institute of Molecular and Cell Biology
Panel: NZ1
Funding scheme
: SYMFONIA 1
announced on
16 December 2013
Mitochondria are structures responsible for many key processes in the living cell, including the production of energy that the cell needs to survive. In people, defects in mitochondria may lead to very serious disorders, such as neurological diseases. A unique feature of mitochondria is that they have their own genetic material (deoxyribonucleic acid, DNA), as well as the machinery to read it. At the first stage, DNA is transcribed into ribonucleic acid, RNA, which is then read during the protein synthesis process. To function properly, mitochondria need an adequate supply of proteins and the main way to ensure their desired quantity is to regulate the levels of protein-coding RNA. This happens primarily through the process of RNA degradation. As a consequence, degradation plays an immensely important role in healthy mitochondrial function.
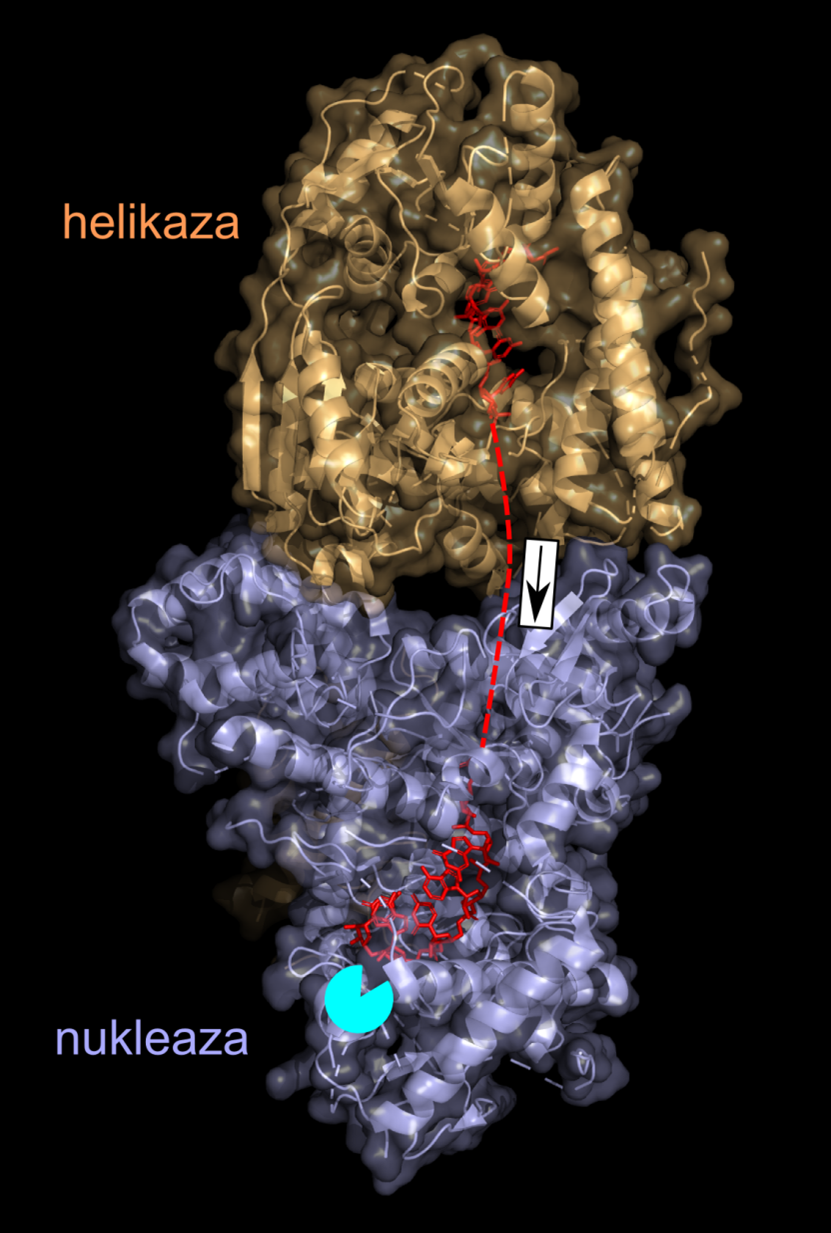 Computer visualisation of the atom's spatial structure in the yeast mitochondrial degradosome. The helicase is marked in orange, nuclease in blue and RNA strand in red. The part not observed directly in the structures is marked with a dashed line. The direction of RNA movement is marked with an arrow, and the place where nucleotides are cut out from the end of the RNA, with a turquoise circle. Author: Marcin Nowotny.
The purpose of our project was to arrive at a comprehensive understanding of the mechanisms of mitochondrial RNA degradation. We wanted to study them both on a more general level, in the context of the mitochondria as a whole, and in much closer detail, on the level of the individual atoms that make up the proteins involved in the process.
Computer visualisation of the atom's spatial structure in the yeast mitochondrial degradosome. The helicase is marked in orange, nuclease in blue and RNA strand in red. The part not observed directly in the structures is marked with a dashed line. The direction of RNA movement is marked with an arrow, and the place where nucleotides are cut out from the end of the RNA, with a turquoise circle. Author: Marcin Nowotny.
The purpose of our project was to arrive at a comprehensive understanding of the mechanisms of mitochondrial RNA degradation. We wanted to study them both on a more general level, in the context of the mitochondria as a whole, and in much closer detail, on the level of the individual atoms that make up the proteins involved in the process.
The project allowed us to identify new proteins that participate in regulating RNA levels. One is the human MTRES1 protein, which plays a protective role, preventing a decrease in functional RNA levels in the mitochondria under harsh conditions. Another protein, known as GRSF1, was shown to cooperate with the degradosome, the main mitochondrial machinery responsible for RNA breakdown. This applies particularly to guanosine-rich RNA sequences (composed of many G letters of the genetic code). Such RNA molecules fold into a very special, stable spatial structures. We demonstrated that the specific role of GRSF1 is to destabilize these structures to facilitate their degradation (article published in “Nature Communications” in 2018). The machinery of the degradosome consists of two proteins. One, known as helicase, moves the RNA chain; the other, nuclease, cuts individual nucleotides (letters of the genetic code) off one end of the chain. To arrive at an in-depth understanding of this process, we used protein crystallography to determine the atomic structure of the yeast degradosome. Helicase and nuclease were shown to bind together to form a compact machinery in which the role of the former is to push the RNA into a tunnel inside the latter. At the end of the nuclease tunnel, there is a site where the chemical reaction of nucleotide excision takes place. Therefore, the two proteins form a strictly coordinated, effective machinery to break down RNA (article published in “Nature Communications” in 2018). We also studied the functioning of the yeast degradosome in more general context and showed that its role is to ensure that functional RNA (including protein-coding RNA) continues to be present in the mitochondria, while non-functional RNA is removed. An important part of the project involved developing computer methods to analyse data on mitochondrial RNA sequences, as well as to examine those RNA regions that interact with proteins. The advantage of these new tools is that they can be employed even with difficult data sets, including those with a great sequence diversity. The project unveiled important information about RNA processing in human mitochondria and the mitochondria of simpler organisms (yeast). This has allowed us to better understand the functioning of these key cell structures. The data on human cells will also provide a basis for understanding a variety of serious disorders associated with mitochondrial defects and creating effective treatment strategies.
Project title: Mitochondrial RNA decay and surveillance - comprehensive interdisciplinary studies
Dr hab. Marcin Nowotny
Since 2008, he has headed a lab at the International Institute for Molecular and Cell Biology. He is a winner of the ERC Starting Grant (2011). Between 2003-2008, he completed a postdoctoral fellowship at the National Institutes of Health, USA. He earned his PhD in 2002 at the Institute of Experimental Biology of the Polish Academy of Sciences.
Team members:
Dr hab. Roman Szczęsny
Since 2017, he has headed a group at the Institute of Biochemistry and Biophysics of the Polish Academy of Sciences. Between 2010-2018, he completed a postdoctoral fellowship at the University of Warsaw and the Institute of Biochemistry and Biophysics of the Polish Academy of Sciences. He earned his PhD in 2009 at the Institute of Biochemistry and Biophysics of the Polish Academy of Sciences.
Prof. Dr hab. Paweł Golik
Head of the Department of Genetics and Biotechnology at the Faculty of Biology of the University of Warsaw. Between 2000 and 2002, he completed a postdoctoral fellowship at Emory University in Atlanta, USA. He earned his PhD in 1999 at the Faculty of Biology of the University of Warsaw, in cooperation with the Centre de Génétique Moléculaire, CNRS in Gif-sur-Yvette, France
Dr hab. Bartosz Wilczyński, Prof. UW
Since 2021, he has been employed as a professor at the Faculty of Mathematics, Computer Science and Mechanics of the University of Warsaw (he became an Assistant Professor in 2011). Between 2008 and 2011, he completed a postdoctoral fellowship at the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany. He earned his PhD in 2008 at the Institute of Mathematics of the Polish Academy of Sciences.
On the photo from left to right: Marcin Nowotny, Bartosz Wilczyński, Roman Szczęsny, Paweł Golik. Photo by Daria Goś
